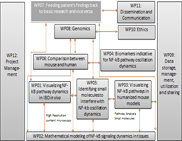
WP05 - Identifying small molecules to interfere with NF-kappa-B oscillation dynamics
Objectives
The aim of this workpackage is to identify potential targets from a predictive regulatory NF-kappa-B network, constructed from literature and new experimental data as well as from upstream analysis of these data (Tasks 1-3). In addition, small molecules potentially interfering with NF-kappa-B pathways will be identified (Tasks 4-5). Finally, the effects of these chemical compounds will be experimentally validated (Task 6).
Workpackage Description
Task 1: Extended NF-kappa-B network
We will analyse publicly available and commercial databases on gene regulation, signal transduction and metabolic networks/pathways of the NF-kappa-B pathway. We will construct an extended gene regulatory and signal transduction pathways using information from those available databases and then extends the pathway through prediction of transcription factor and miRNA targets and by reconstructing upstream signalling cascades. Novel methods for high quality prediction of functionally important TF binding sites will be implemented in the geneXplain platform through statistical analysis of TF site combinations by integrating knowledge on available gene expression data and large scale ChIP-seq data and epigenomic data. The resulting network will be compared and merged with the one generated by Lifeglimmer obtained by multivariate statistics and reverse engineering procedures to the trancriptomics and proteomics data generated in the consortium.
Task 2: Integrated tool platform
To enable use of omics data for identification of targets and candidate molecules against these targets, the corresponding tools will be integrated in the geneXplain platform.
Task 3: Extend the training set for PASS and GUSAR for ligands of the selected targets
Currently, the SAR base of PASS contains a training set of 1,307 currently used drugs and known compounds implicated in treatment of IBD. The overall size of the training set of PASS exceeds 250,000 compounds associated with more than 5,000 different biological activities and various molecular targets. Those that may be relevant for project goals will be selected and added to the SAR base. PASS will be re-trained accordingly.
Task 4: Small molecules interfering with NF-kappa-B pathway
Applying the PASS and GUSAR software, a series of small molecules will be predicted that are supposed to interfere with components of the NF-kappa-B pathway. The effects of these and other suitable molecules on the network structure and network dynamics will be simulated.
Task 5: Protein target prediction methodology and application to NF-kappa-B network
The extended NF-kappa-B network constructed under Task 1 as well as networks that are constructed in the consortium will be analyzed for key molecules as potential targets. Existing topological and statistical methods for such an analysis will be refined, adapted to the specific project requirements and applied. Results from the experimental validations
(Task 6) will be used to refine the network and the target predictions.
Task 6: Experimental validation of selected small molecules interfering with NF-kappa-B pathway dynamics
Selected small molecules will be tested in vitro in an established neuroblastmoma cell line for their potential to interfere with NF-kappa-B pathway dynamics. A further selection of compounds will subsequently be validated through in vivo experiments.