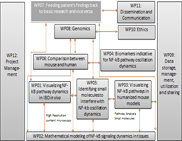
WP06 - Comparison between mouse and human
Objectives
The aim of WP06 is to develop a coherent framework that incorporates experimental evidence from literature sources as well as the model systems used within SysmedIBD. to translate NF-kappa-B findings from mice to humans. The specific objectives are:
- Build semantic tools to automatically translate findings related to NF-kappa-B signalling from mouse to human (Task 1)
- Identify differences between both species and to account for these in the underpinning mathematical models (Task 2)
- Define the best minimal model that incorporates information from human, mouse and chimeric systems (Task 3).
Workpackage Description
Understanding how cells respond to inflammatory inputs such as cytokines or molecules derived from infectious microorganisms is fundamental to our ability to treat chronic inflammatory diseases such as IBD. Reflecting the importance of the NF-kappa-B signalling network in regulating inflammation, the literature holds many thousands of scientific publications, each of which hold potentially valuable information. Mice are very commonly used as models to understand human disease states, also in inflammation and IBD-related diseases. It is therefore crucial to understand how details of physiology in the two systems are related and how coding phenotype data can be meaningfully translated. Hence, the focus of WP06 is to define critical similarities and differences in the murine and human NF-kappa-B signalling network and so establish which parts of the mathematical models are common in the two species. This is a complex endeavour as individual cell types are also likely to display differences in the network architecture, owing to the very complex nature of the network structure and cell type specific differences in expression of components. This is especially relevant at the level of signalling inputs as different cell types that respond to NF-kappa-B can have quite different expression of the various membrane receptors that are able to induce NF-kappa-B signalling.
Task 1: Develop a coherent logic for extracting evidence from text sources to populate models
A direct, automatic translation of phenotype findings from mice to humans is currently not possible as such due to the very different ontologies used for both mammals. Whereas mouse ontologies are well defined and structured, those for humans tend to be vague and include clinical observations that, albeit valuable, are not standardized enough for a rigorous ontology. Indeed, a critical issue in extracting information from texts is the variability in the way in which terms are used in human and mouse systems and particularly difficulties in relating disease states with classical phenotypes ? which are used more commonly in mouse genetics.
This paper has a deterministic model which uses essentially human data and a stochastic model that also has various parameters derived from mouse.
As the cycling activity is innate it is reasonable to assume that it operates similarly in animals. [For NF-kappa-B this has not been formally shown but we do know that the circadian clock which has the same sort of oscillatory behaviour and also p53 both work in cell culture as they do in animals. Also tissues explanted from animals show immediate oscillatory behaviour at the level of DNA association (Hoffmann-A) genome-wide changes in expression (Iqbal-J)]
Task 2: Establish a modelling framework for comparing NF-kappa-B signalling in mouse and human cells
SysmedIBD Partners 01UNIMAN and 04WARWICK have built the, thus far, the most advanced models of the NF-kappa-B signalling network architecture. These models rely on known parameters of the signalling system (e.g. protein concentrations and rate constants) and focus on the way in which pathway induction allows the NF-kappa-B transcription factors to release from the I-kappa-B complexes in the cytoplasm and then move to the nucleus and activate target gene expression . As the behaviour of the model is central to the thorough analysis of the signalling network in WP3 it is critical to be able to assess similarities and differences in the way the pathways operate and if/how the signalling response varies between cell types. The oscillatory behaviour observed was shown to be a stable and robust feature of the system in mouse and human cells. This joint modelling framework combines both deterministic and stochastic models, and both rely on parameters from derived from mouse and humans. We will thus build upon this framework to develop a modular platform to incorporate new data, both curated from literature and produced within the project. We will expand modularity and comprehensiveness to account for the NF-kappa-B related features.
Task 3: Define the best minimal model that incorporates information from human, mouse systems.
We will use mouse models as our basic experimental in vivo tool (WP01 and 03). We have to be able to translate these findings to clinical findings in humans (WP07). The work plan is to build semantic translation tools, to automatically translate findings on the fly between mouse and human (09LIFEGLIMMER).
By experimentally comparing the behaviour of human and mouse cell culture models with the mouse/human chimeric model, we will be able to identify any species differences and account for the differences in the mathematical models. The deliverable of WP06 is therefore to combine findings in mouse and humans into a database language that is able to compare and to compute data from both species. Of course these experiments rely on data generated by time-lapse imaging of living cells with ectopically expressed NF-kappa-B-fusion proteins (usually fluorescent but sometimes luminescent). This live cell output should also be related to analysis of fixed samples in WP04.