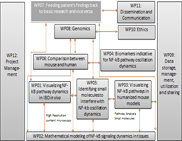
WP09 - Data storage, management, utilisation and sharing
Objectives
WP09 will develop and administer the data management platforms and will lead the data analysis tasks of the project developing new tools, pipelines and workflows as needed
- Provide the environment and tools for data management. sharing and utilisation (Task 1).
- Facilitate the analysis of genomic and imaging data arising from the other WPs (Task 2).
- Facilitate the linking of models and data and the use of models for experimental design and hypothesis generation (Task 3).
Workpackage Description
Data management sharing and utilisation. A key objective is to define and setup a data management policy in order to ensure optimal storage, management, use and sharing of data. In that context, the key requirements are: (i) systematic archiving of data and associated metadata; (ii) integration of results from different experimental techniques; (iii) systematization of the analysis procedures to ensure reproducibility of results; (iv) collection and dissemination of the key facts that are used in the construction and validation of SB models and (v) timely public access to the raw data and principal results. WSB and geneXplain have extensive prior experience of data management in large SB projects (such as, for WSB, the BBSRC funded SABR PRESTA, ROBust and NF-kappa-B projects, the ERASysBio+ C5Sys project and for geneXplain, FP6 projects Net2Drug, SysCo and FP7 projects: LipidomicNet and SysCol). A simple structure based on four core components has been chosen. These include (1) a Wiki, (2) the SysMo DB platform, (3) geneXplain platform, (4) a specialized data web portal for SysmedIBD members. In addition, it is expected that WSB and the White group will jointly implement the OME system Omero in order to handle the project images in a secure central repository. The project will exploit the data management platforms already in current use within WSB and the project partners, with WSB providing linking services (e.g. web services) where needed. Published biological models will be submitted to the EBI?s BioModels database.
Project Wiki. Current successful project Wikis will be cloned and put in place at the start of the project (http://www.wsbc.warwick.ac.uk/twiki/). The wiki will be regularly populated by group members with internal or external key facts relevant to the project. Topics include for instance, raw and processed data, models, gene lists, SOPs and reporting (see capture below). Main advantages are easy access and editing, flexibility, interactivity and possibility to link to external databases. The primary goal of the wiki is to stimulate the sharing of data and information in the early stage of the development of the different part of the project before they become relevant to other consortia through SysMo DB.
SysMoDB platform. SysMo DB is already in place within WSB (04 UNIWARWICK) who have used it extensively. It will be available from day one with a dedicated team that can assist new users (led by of Jay Moore an experienced WSB data manager and SysMo-DB PAL). It compares favourably with other solutions (basic front end or wiki for instance). All partners have agreed to use the SysMO DB platform for storing and sharing data within the project.
GeneXplain platform for data analysis. The geneXplain platform (![]() http://genexplain.com/genexplain-platform/) is an online toolbox and workflow management system for a broad range of bioinformatics and systems biology applications. The platform is based on the open source BioUML systems biology plug-in based framework
http://genexplain.com/genexplain-platform/) is an online toolbox and workflow management system for a broad range of bioinformatics and systems biology applications. The platform is based on the open source BioUML systems biology plug-in based framework
and will be provided to the project partners. The individual modules, or Bricks, are unified under a standardized interface, with a consistent look-and-feel and can flexibly be put together to comprehensive workflows. The workflow management is intuitively handled through a simple drag-and-drop system. With this system, user can edit the predefined workflows/pipelines or compose your own workflows from scratch providing the standard analysis procedures for data analysis in the SysmedIBD project. New analysis methods ? new Bricks, developed by 10 GENEXPLAIN and other partners of the project will be added as scripts (R, Java script, Perl, Python, etc.) or Java or C++ plug-ins and can be used in combination with pre-existing analyses tools. The geneXplain platform will be integrated with SysMo DB to enable access to the consortium data for analysis pipelines.
Data web portal. The primary goal is to enable easy data transfer and manipulation, access to software/algorithms (e.g. for imaging, network reconstruction, bioinformatics and timeseries analysis tools) and use of web services.
Genomics analysis. A broad range of tools will be available for the analysis of genomics data such as microarrays, nanostring, and second generation sequencing. In addition to preparation and normalisation these include tools for identifying differential expression, hierarchical clustering and clustering of heterogeneous data, network reconstruction, promoter analysis and transcription factor binding site prediction. Pipelines are available for the analysis of RNA-seq and ChIP-seq data. WSB and geneXplain staff are experienced in using all these aspects.
Image analysis. WSB will provide access to the extensive research experience in WSB?s Bretchneider group. This include software for (i) tracking nuclear and cytoplasmic fluorescence intensities from live cell microscopy time series data (CellTracker), (ii) quantification of spatio-temporal patterns of fluorescently labelled proteins in the cortex of moving cells (Quip), and (iii) tracking cell lineages while being specifically designed to handle large cell displacements between frames (Lineage Tracker).
Linking imaging time series to models. WSB The current broad range of statistical techniques developed and used by WSB will be applied with 03DKFZ to the experimental time-series generated in the other WPs (e.g. the luminescent and fluorescent measurements from WPs 1, 3 & 5) in order to parameterise and validate key aspects of the models.
Model analysis & experimental optimisation and navigation WSB will provide detailed analysis of NF-kappa-B and other models and the interaction between them to determine their robustness and sensitivity, to identify key nodes of interaction for these networks and to understand their design principles (17, 20, 51). These tools will be used to tune and modify the ODE and stochastic clock and other oscillatory models to incorporate new data from mutant or perturbation experiments such as coming from 09LIFEGLIMMER(WP06). WSB will also use this and related experimental navigation software to guide the choice of next experiments allowing a more rational/mathematical approach to planning and optimisation of experiments that can integrate multiple & heterogeneous data sources.
Task 1: Sysmo-DB database, geneXplain platform, wiki and webpages in place (months 1-12).
Task 2: Workflows/pipelines for image and genomics analysis in place (months 1-36) and developed to meet need
of project (months 1-60).
Task 3: Pipelines for model analysis, linking models to data and experimental design in place (month 1-36) an further developed to meet needs of project (months 1-60).